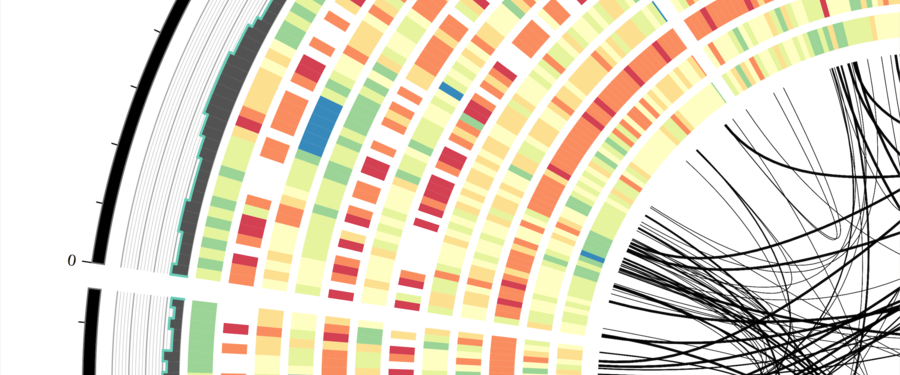
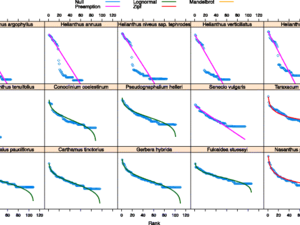
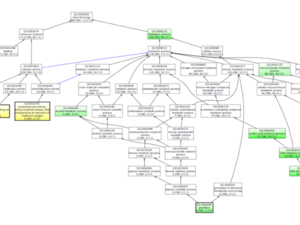
Professionally, I am a computational biologist and application developer that is passionate about writing quality software to improve science and facilitate discovery. My background is in biology with a focus on plants. I am trained as a plant geneticist and most of my work centers around developing novel computational methods for understanding patterns of genome evolution.
I am currently employed with a private agricultural genomics company in Saskatoon, SK, Canada where I work on delivering insights for a wide variety of projects. Formerly, I was a Research Associate at the Beaty Biodiversity Research Centre and Department of Botany at UBC. If you would like to discuss one of my projects or a collaboration, please send me message.
When I am not working I enjoy staying active (cycling, hiking, running, rock climbing) and above all I things I enjoy reading to learn new things.